Projects
Few of my research projects during my Ph.D. and postdoc are listed below.
My research experience can be divided into two distinct groups: (1) developing generic Active Learning (AL) methods and applying them on various applications including medical image processing, and (2) applying machine learning and network analysis to stuy different aspects of knowledge production and scientific discoveries.
Contents
Active Learning
Active learning (AL) is a branch of machine learning that addresses the problem of minimizing the cost of data annotations necessary for training accurate learning models. This task is increasingly paid more attention in the era of data science where raw, unannotated data are available in low cost but their labeling is time-consuming and financially expensive. Classically, AL methods assumed a fixed budget for labeling samples, and formulated the problem as how to select a certain number of queries such that their annotations will likely provide the most amount of information towards the best model fit.
Generic AL
A major part of my research included addressing two shortcomings of the most popular querying metric in AL, that is entropy-basede Unertainty Sampling (US). This metric selects those queries about which the model has the highest uncertainty. While US has a sound mathematical optimization problem, in practice it often leads to selecting redundant queries and/or outliers. These types of queries are not desirable in AL settings since they do not provide maximum information gain. The main source of sub-optimality in entropy-based US goes back to considering individual labeling candidates separately in the querying optimization.
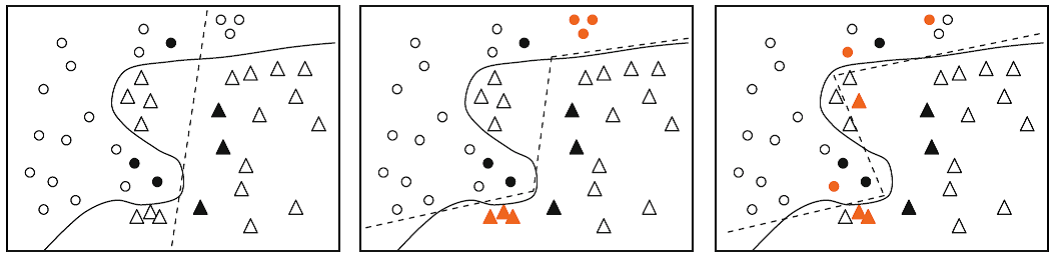
Numerous hueristics have been suggested to fix these issues of US. We proposed to change the querying objective to a more sophisticated information theoretical metric, i.e., Mutual Information (MI) and Fisher Information (FI). Both MI and FI by definition have mechansims that enable them to automatically address above issues: MI measures expected information gain of all the candidate queries and their (unknown) annotations collectively in its formulation, and FI aims at choosing samples that are most informative regarding the joint distribution of the whole data.
Our MI- and FI-based algorithms were first of their own kind. Our MI framework attacked the querying problem via directly computing MI between discrete class labels of the query candidates despite all the previous attempts that worked with crude approximations. On FI’s side, we theoretically explained how FI-based active learning can improve the information gain when being used in conjunction with maximul-likeliood estimators. Our derivations have been published in the Journal of Machine Learning Research (JMLR) (see here), where we reviewed other similar methods and demonstrated their theoretical drawbacks with regards to ours.
More recently, I have collaborated with a brilliant team to analyze practical settings of AL algorithms and shed lights on how various experimental settings could affect performance of different AL techniques. The outcome of this analysis together with a set of proposed guidlides for facilitating a more robust and reproducible AL framework will be published shortly in CVPR 2022 (see the preprint here).
Application to Medical Image Processing
Our proposed information theoretical AL methods explained above dramatically outperformed entropy-based US and random sampling baselines on popular machine learning benchmarks (e.g., UCI datasets). Testing them on a real-world application was whta I did in my first postdoc at Boston Children’s Hospital, Harvard Medical School.
In hospitals gigantic amount of unlabeled images are being produced on a daily basis and there is a need for AI-assisted algorithms to increase the speed and accuracy of their interpretation. There are several impediments in doing this task. Here’s a brief list:
- The pre-trained state-of-the-art deep learning models cannot be readily applied to all groups of patients due to large visual characteristics of their medical images.
- Training or fine-tuning deep models is not possible for many patient groups as enough annotated data is not available for all ages or pathological conditions.
- Preparing annotations for medical images is a time-consuming and costly task and in most cases only a small number of image volumes can be labeled due to budget considerations.
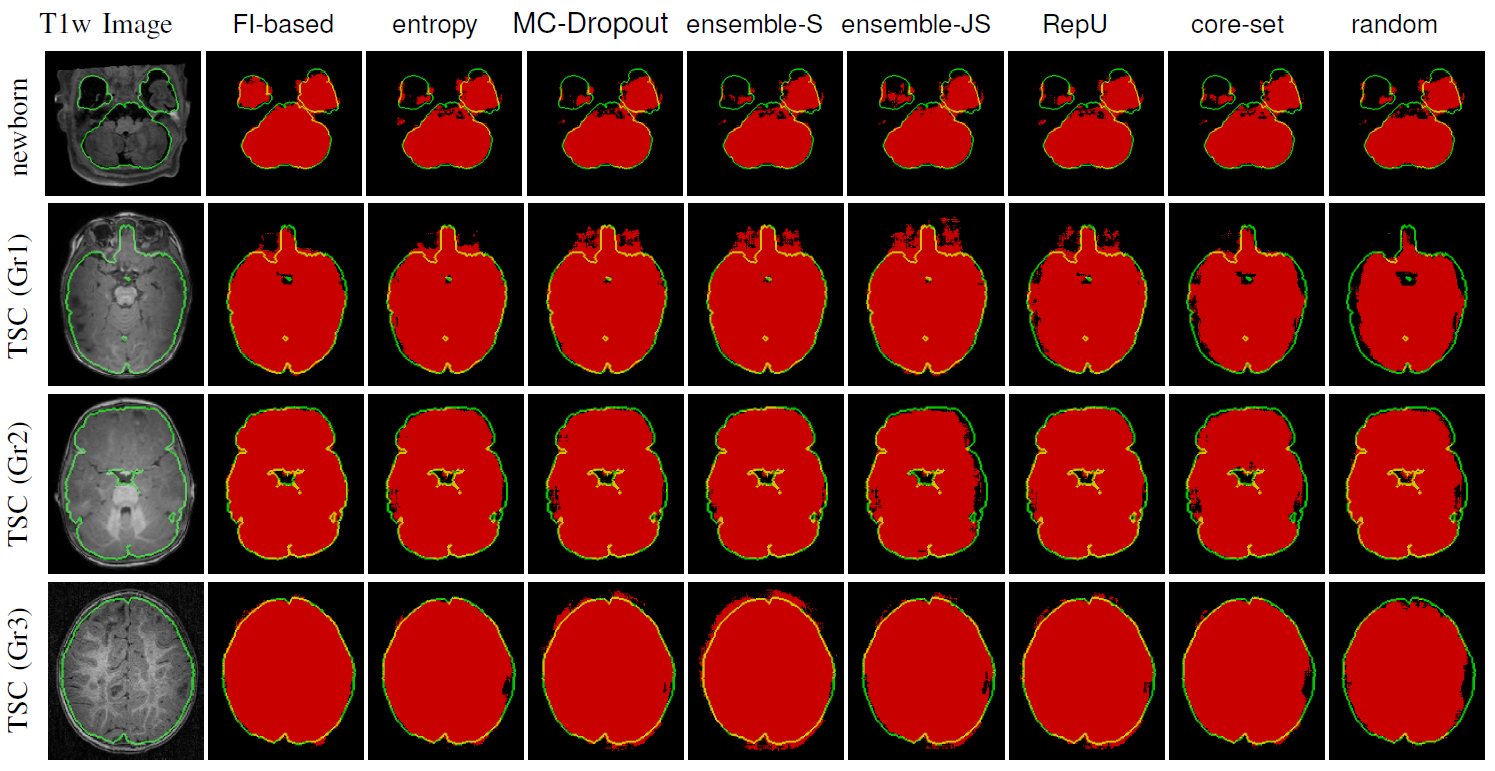
These issues sound like a good application of AL to train or fine-tune a deep model with a fixed labeling budget. We slightly modified our FI-based AL to make it tractable for deep models and applied it with Convolutional Nneural Networks (CNN) on the problem of IntraCranial Cavity (ICC) extraction in brain MRI images. We worked with three distinct patient groups: healthy adolescents, newborns (with age between two months and 2.5 years) and patients with Tuberous Sclerosis Complex (TSC)}. Visual characteristics of these images, specifically T1- and T2-weighted MR images, vary extensively among these groups. For example, we noticed that T1-MRI images of newborns show a completely reversed intensity contrast compared to older subjects.
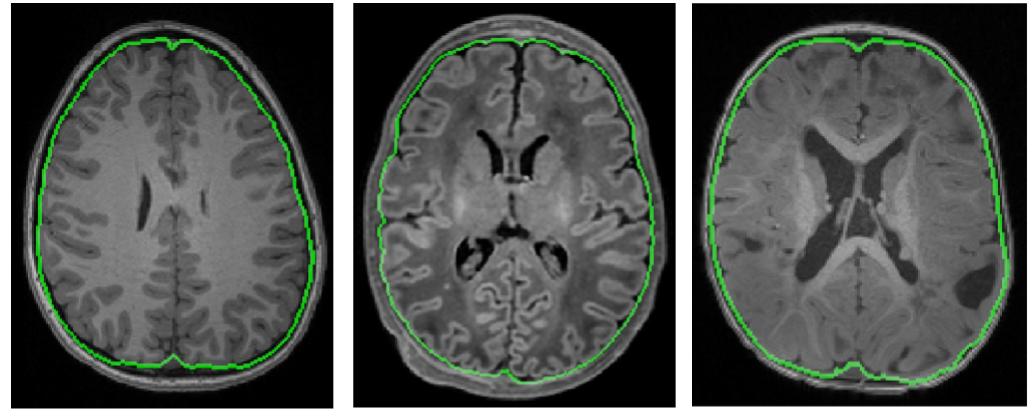
Because of major variabilities across the images, models trained on one group will not be accurately generalized to the rest. We pre-trained a CNN based on healthy adolescents and used our modified FI-based AL to fine-tune it to the other two groups with limited number of annotations. Comparing the results with multiple variations of US that were common in medical image processing indicated that using FI enabled the model to reach a high performance with significantly smaller number of annotaions. In the setting explained above, this means that using FI queyring metric lead to a more generalizable model with a fixed labeling budget.
Interactive Unsupervised Learning
All I explained so far was the development and application of AL in supervised learning problems. I also worked with similar issues in the context of unsupervised learning. Indeed, this line of research formed the very first project that I did in my Ph.D.. Since AL comes to the scene when some kinds of interaction exists between the machine and the user, we considered constrained clustering algorithm where the user-provided pairwise constraints were in form of must- and cannot-links.
The idea of entropy-based US can also be applied in this problem, but with the same redundancy and outlier vulnaribility issues. In my project, we addressed the latter issue by combining entropy and an estimate of data density to make sure that the queries are located in dense regions. We applied our density-weighted AL algorithm with an extended version of spectral clustering that could be scaled to segment intermediate size to large images. The final product was a practical user-interactive unsupervised framework that could segment complicated images by only using naive intensity features and with the help of AL-accelerated user pairwise constraints.
AI-assisted Knowledge Discovery
My personal interests in humanities and social sciences drove me to start a second post-doc at Knowledge Lab in the Sociology Department of University of Chicago. In there, supervised by prominent computational sociologist James A. Evans, I becamse familiar with the topic of Science of Science and started working on development and deployment of computational tools to study how science advance by training word embedding and language models over the vast existing literature and the underlying authorship network, which also enabled us to build predictive models to accelerate knowledge discoveries.
Expert-aware AI for Accelerating Future Discoveries
We investigated the importance of incorporating authors and their interconnections in the computational modeling of scientific findings. Artificial intelligence (AI) algorithms have been developed over scientific publications to automatically identify discovery candidates from the vast space of all possible combinatorial scientific knowledge. As an example, a work by Tshitoyan et al. (2019) utilized skipgram word2vec model to predict which materials possess electrochemical properties such as thermoelectricity. By training a word2vec embedding model over materials science literature, they could achieve a precision of %40 in predicting future thermoelectric discoveries occurring in two decades. This work and other similar algorithms neglect authors of the publications that they used to create their predictive models. Our idea was to incorporate the distribution of scientists and experts, the main workforce of science’s engine, in computational analysis of advancement of scinece.
The intuition is that the underlying links between scientific topics could be discovered only when the corresponding scientific communities start to communicate. For instance, if material X (e.g., germanium telluride) does show property Y (e.g., thermoelectric effects) in reality, the link X-Y will not be discovered as long as there is no bridges between the communities around X and Y. The prelude of any realistic discovery is that, say, a crowd of collaborators, some of which have expertise in X and some know Y very well, communicate with each other to put forth hypotheses regarding X-Y relationship and experiment them. Such a communication between these experts makes X-Y relationship cognitively availabile to other scientists, which is key to discovering scietific knowledge.
In order to incorporate expert distribution into our model, we built a hypergraph that included both author nodes and conceptual nodes (e.g., materials and their properties). The hyperedges comprised of papers linking nodes based on the authorship and mention of the materials/properties in their title or abstract. We then predict the future hypotheses by pairing material-property nodes with the largest hypergraph-induced similarities, which could be measured by any graph representation algorithm such as deepwalk embeddig.
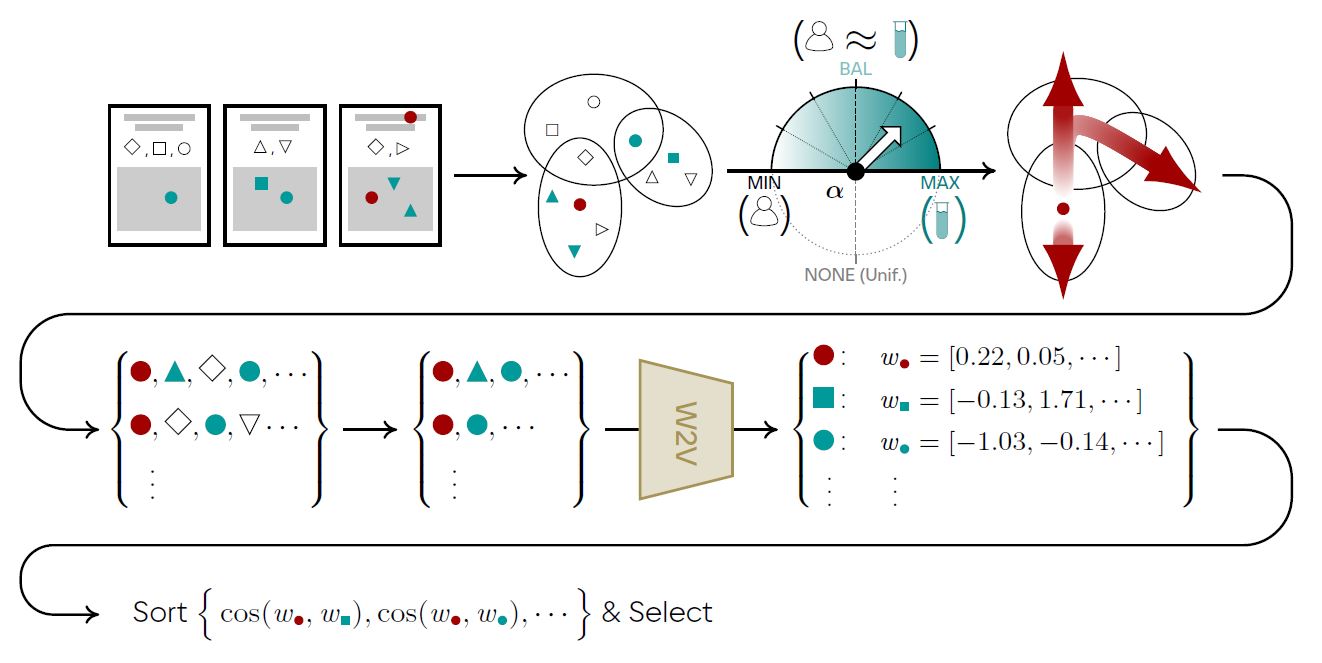
We evaluated the effect of introducing experts distribution on the quality of discovery predictions in the context of materials science, disease therapies and vaccination. Numerical results showed that applying our algorithm led up to 100% precision increase in distinguishing materials with certain enery-related properties (thermoelectricity, ferroelectricity, being photovoltaic), average 43% increase in drugs repurposing for one hundred human diseases, and a boost of 260% in therapy/vaccine prediction for COVID-19 (see the figure below).
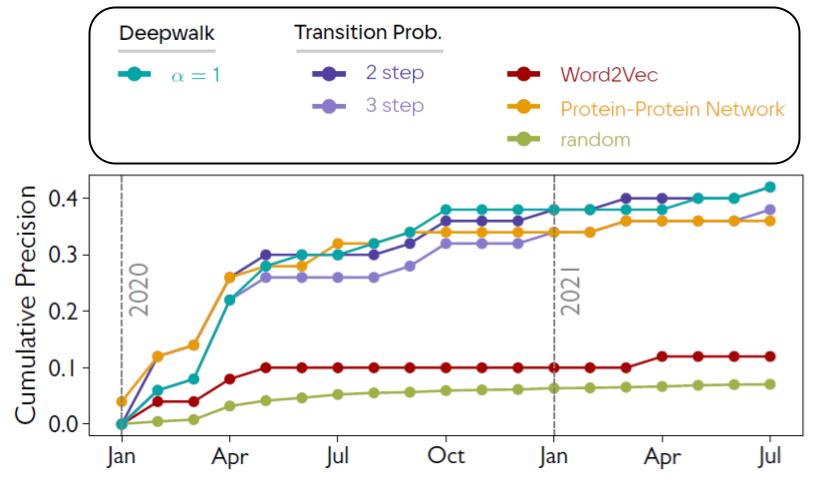
Complementary AI for Punctuating Disruptive Discoveries
Our expert-aware algorithm described above also enabled us to avoid the distribution of experts to generate hypotheses that are not imaginable by human scientists without machine intervention. In other words, here, we look for unbridged communities for which their corresponding topics seem to have an undiscovered scientific relationship. Since there is no communication between these communities, any relationship between their topics is very likely to be cognitively unavailable and, therefore, is likely to escape scientists’ collective attention. Historically, this was the aim of Don Swanson when he manually searched for unlinked scientific topics that were potentially related. For example, he noticed tha Raynaud’s disorder and fish oil were both somehow linked to blood viscosity, consequently he conjectured a relationship between fish oil and Raynaud’s disorder, a relationship that could not be organically discovered as there was no scientist available to bridge the communities and infer the relationship. This hypotheses was later experimentally demonstrated by experts of the fields (see more details here).
Our algorithm scales Swanson’s approach and makes it continuous by explicitly measuring the distribution of experts as well as drawing upon advances in word embedding models. We developed a simple algorithm to identify materials that are far from the property node in our hypergraphmentioned above (in terms of shortest-path distance) and at the same time show high semantic relationship with the property. These materials are coginitively unavailable to scientists and authors due to their high distance to the property, yet are potentially good candidates of possessing the property due to their high semantic similarities with respect to the property. We showed that in practice such “alien” hypotheses could be very promising even when they are infinitely far from the property node (see the figure below).
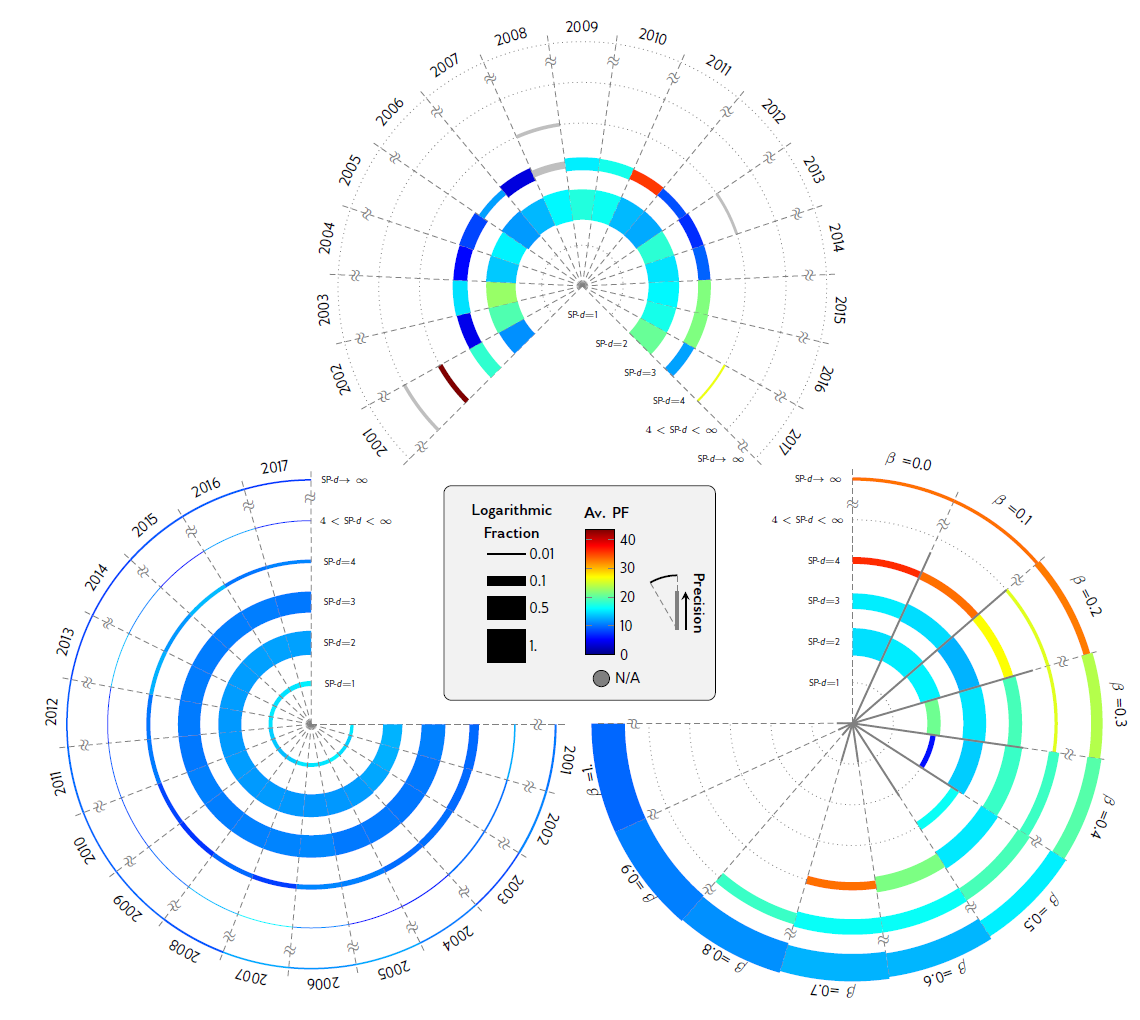
Our findings imply that the pattern of scientific discoveries is highly predictable merely through the coauthorship network and the prior experience of the scientists, without any prior knowledge of the underlying theories and even without the knowledge of the content of the prior literature. These patterns, in one hand, could enable us to predict the upcoming future discoveries, and on the other hand, help us distinguish and escape collective biases in the course of scientific discoveries, and thereby open doors to previously unimaginable hypotheses.
Measuring Language Uncertainty in Scientific Communications
There is always a degree of uncertainy lying within each scientific findings and this uncertainty could be reflected in the scientific writings when communicating the results. It will be quite useful to be able to quantify this uncertainty and incorporate it when automatically parsing the literature. In this ongoing project, we employ several methods for computing the language uncertainty in publications of various scientific fields and study their relationship with the objective uncertainty of the findings. We will also investigate possible patterns of usage of language uncertainty in various scientific fields and in different periods of time.
One challenge of this task is the scarcity of relevant annotated datasets. Each of the individual datasets usually have small number of annotated statements, in which “certain” statements highly outnumber. We also argue that even the human annotations are biased towards the very certain and/or very uncertain extrema mislabeling neutral expressions. Here, we create an ensemble-based approach for measuring the language uncertainty with a hope to lessen the bias in each individual model. Our ensemble includes
- an LSTM trained from scratch over a set of uncertainty-annotated statements (provided by this paper),
- a zero-shot model based on a pre-trained large-BART transformer, in which we computed normalized scores indicating the relevance of given statements to tokens “certainty” and “uncertainty” (pre-trained model available in huggingface.co),
- a SciBERT model fine-tuned over uncertainty annotations by Pei and Jurgens (2021),
- a hedge-based approach that scores the uncertainty of any given statement based on the number of mentions of a list of hedges–this is the most traditional method of the ensemble.